| << Chapter < Page | Chapter >> Page > |
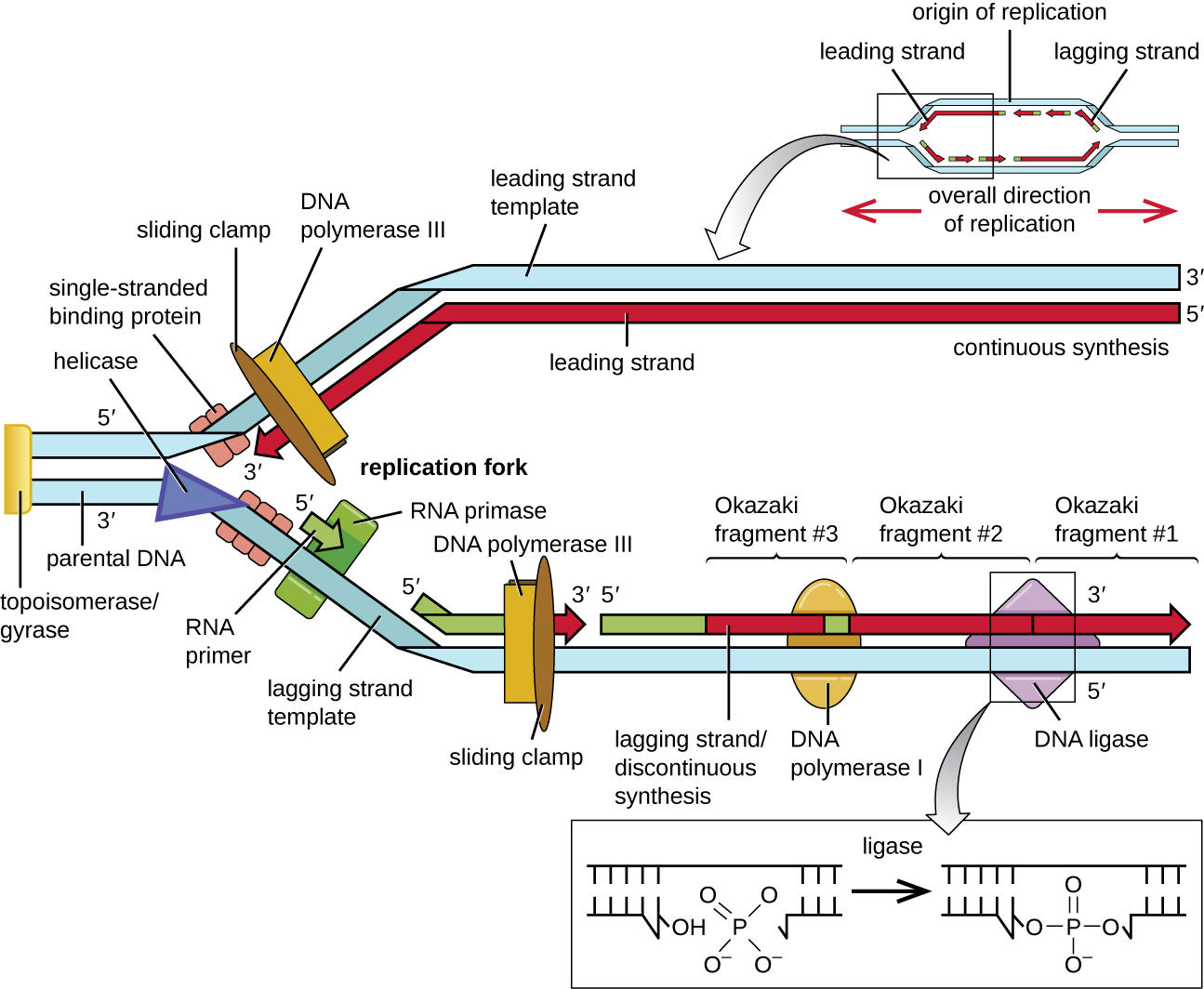
Once the complete chromosome has been replicated, termination of DNA replication must occur. Although much is known about initiation of replication, less is known about the termination process. Following replication, the resulting complete circular genomes of prokaryotes are concatenated, meaning that the circular DNA chromosomes are interlocked and must be separated from each other. This is accomplished through the activity of bacterial topoisomerase IV, which introduces double-stranded breaks into DNA molecules, allowing them to separate from each other; the enzyme then reseals the circular chromosomes. The resolution of concatemers is an issue unique to prokaryotic DNA replication because of their circular chromosomes. Because both bacterial DNA gyrase and topoisomerase IV are distinct from their eukaryotic counterparts, these enzymes serve as targets for a class of antimicrobial drugs called quinolones .
| The Molecular Machinery Involved in Bacterial DNA Replication | |
|---|---|
| Enzyme or Factor | Function |
| DNA pol I | Exonuclease activity removes RNA primer and replaces it with newly synthesized DNA |
| DNA pol III | Main enzyme that adds nucleotides in the 5’ to 3’ direction |
| Helicase | Opens the DNA helix by breaking hydrogen bonds between the nitrogenous bases |
| Ligase | Seals the gaps between the Okazaki fragments on the lagging strand to create one continuous DNA strand |
| Primase | Synthesizes RNA primers needed to start replication |
| Single-stranded binding proteins | Bind to single-stranded DNA to prevent hydrogen bonding between DNA strands, reforming double-stranded DNA |
| Sliding clamp | Helps hold DNA pol III in place when nucleotides are being added |
| Topoisomerase II (DNA gyrase) | Relaxes supercoiled chromosome to make DNA more accessible for the initiation of replication; helps relieve the stress on DNA when unwinding, by causing breaks and then resealing the DNA |
| Topoisomerase IV | Introduces single-stranded break into concatenated chromosomes to release them from each other, and then reseals the DNA |

Notification Switch
Would you like to follow the 'Microbiology' conversation and receive update notifications?